Final Report
Group #13 - Kezi Cheng, Nikhil Mallareddy, Jonathan Fisher
1. Problem Statement and Motivation:
Alzheimer’s Disease (AD) is a common and devastating neurologic disease. Although the pathologic features of AD are easily recognizable on invasive brain biopsy, no other single clinical test has proven its utility as a gold standard for AD diagnosis, and standard of care still relies on clinical acumen. The development of better standardized clinical tests that in combination could accurately diagnose AD would greatly reduce the need for intensive clinician involvement in diagnosis, freeing up neurologists for other tasks. The aim of our analysis is to develop a clinical prediction tool that can closely analyze the effect of different bundles of diagnostic predictors on final prediction accuracy of AD. We will try to identify the minimal set of such predictors that can lower the overall cost of clinical testing without sacrificing a substantial degree of diagnostic accuracy.
2. Introduction and Description of Data:
We have chosen to focus our analysis on the ADNIMERGE and UPENN CSF datasets. The ADNIMERGE dataset contains data from 1784 unique patients across multiple ADNI protocols and several dozen of the most widely studied clinical variables relevant to dementia. These include demographic data such as age, race, and gender; cognitive assessment scores from tests such as CDR and MMSE; ApoE genotype; and neuroimaging analyses such as average PET signals and estimated brain volumes from MRI. The outcome of interest is dementia diagnosis in one of three categories: normal (CN), possible/subclinical dementia (MCI), and dementia (AD) (the original DX_bl column contains diagnoses in five categories which were collapsed into these three as follows: CN+SMC=CN, LMCI+EMCI=MCI, dementia=AD). Our analysis focuses only on data collected at each patient’s baseline visit in order to simplify interpretation and to build a model that can predict disease status based on a single clinical snapshot without requiring follow-up visits, a significant barrier to timely and efficient diagnosis.
The UPENN CSF dataset adds additional information on the measurements of key neuro-substances implicated in AD and found in the cerebrospinal fluid (amyloid beta, TAU, and p-TAU). These CSF data are available for 1238 of the patients in ADNIMERGE. Although not yet adopted in standard practice, these measurements have been shown to contain predictive value in AD diagnosis. In initial cleaning of the CSF dataset, we chose to use ‘median’ values which represent a summary of CSF measurements across multiple batches. In combining this CSF data with ADNIMERGE, we merged on patient ID (RID) for all baseline observations in each dataset.
ADNI which stands for “Alzheimer’s Disease Neuroimaging Initiative” is a multisite study that aims to improve clinical trials for prevention and treatment of Alzheimer’s Disease. The study consists of four cohorts, ADNI-1 which lasted from 2004-2009, ADNI-Go from 2009-2011, ADNI-2 from 2011-2016, and the ongoing ADNI-3 from 2016-2021. Each subsequent cohort has a variety of primary goals that varies from developing biomarkers as outcome measures for clinical trial, examining biomarkers in earlier stages of disease, to the use of tau PET and functional imaging techniques and clinical trials.
From the ADNIMERGE dataset, we decided to focus our analyses on only the ADNI 2 dataset for several reasons. Primarily, ADNI-2 contains additional metrics from MRI as compared to ADNI-1, yet it contains almost the same number of participants as ADNI-1. Secondly, when we looked at the missing values for ADNI-1, we found that over 50% of the participants were missing 8-15 predictors, mainly because many of the participants in ADNI-1 did not have corresponding CSF data. To impute all of these values from the ADNI-1 data would give us an unrealistic dataset.
3. Literature Review/Related Work:
The literature and online sources were consulted for aid in estimating costs associated with clinical tests. Healthcare Bluebook (www.healthcarebluebook.com) provided average estimates for costs of psychological testing ($192) and brain MRI with contrast ($1102). New Choice Health (www.newchoicehealth.com) was used to estimate the cost of a PET scan brain ($7700), and we considered this cost to cover both FDG-PET and AV-45 simultaneously, since this involves only one additional sequence in imaging acquisition. The cost of a home ApoE genetic test was found to be $149 (www.lifeextension.com/Vitamins-Supplements/itemLC100059/APOE-Genetic-Test-for-Alzheimers-and-Cardiac-Risk). Costs of cerebrospinal fluid analysis were estimated at $463 in a previous study (Lee SA, Sposato LA, Hachinski V, Cipriano LE. Cost-effectiveness of cerebrospinal biomarkers for the diagnosis of Alzheimer's disease. Alzheimers Res Ther. 2017;9(1):18.), and this cost was assumed to cover the three substance measurements in our analysis. Demographic information was considered to have zero cost because it can be obtained directly from the patient.
We also considered the monetary savings associated with detecting Alzheimer’s earlier versus later. Barnett et al. estimate that detecting Alzheimer’s ten years earlier results in an average savings to the healthcare system of $5508 due to direct costs of disease management (Early intervention in Alzheimer's disease: a health economic study of the effects of diagnostic timing. BMC Neurol. 2014;14:101). Furthermore, indirect costs such as rehabilitation and living assistance averaging $6350 are saved by detecting disease ten years earlier. We therefore consider these total savings of $11858 to be the savings associated with early Alzheimer’s detection by a clinical test. We categorized the clinical tests in our analysis according to their ability to detect disease early, medium, or late, with respect to disease onset. The only test considered ‘early’ is genetic testing, which can theoretically be done at or before birth. Demographic factors, some of which are static but others of which may change (such as age) were considered overall ‘medium’, and CSF biomarker measurement was also categorized as ‘medium’ since biochemical changes in the CSF biologically precede structural brain changes as detectable on neuroimaging as well as clinical changes as evident on neuropsychological testing. PET, MRI, and neuropsychological tests were all considered ‘late’ detectors of Alzheimer’s.
4. Modeling Approach and Project Trajectory:
After successfully merging the ADNIMERGE and CSF datasets, mean imputations were performed on features with quantitative values, and mode imputations were performed on features with categorical values. Furthermore, one hot encoding was performed on categorical predictors and baseline predictors were dropped due to redundant information. Standardization was performed on the feature sets with continuous data. This leaves a final cleaned dataset where a 75% to 25% train-test split was performed. The train dataset was further split into 80% to 20% train-validation.
Six classification baseline models were used to train the clean dataframe and scored using model classification (prediction) accuracy. Multinomial logistic model was chosen for further feature selection due to relative familiarity and model interpretability. Backward selection was performed to find a minimal predictor set with greatest predictive power.
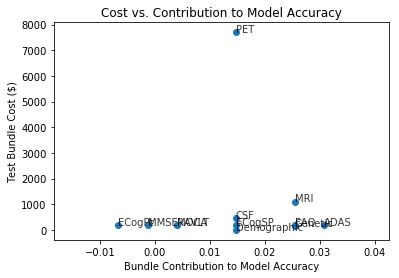
Features were bundled according to assessment of shared costs. For example, all MRI metrics are assumed to result from a single brain MRI scan with a baseline cost, and therefore we consider all MRI metrics as a predictor bundle. Each neuropsychological test is considered as an individual one-predictor bundle since the main cost of neuropsych testing is a professional’s time, and each additional test costs additional time. Direct costs of tests and indirect cost associated with detection timing were estimated as described above in Literature Review. In order to assess the predictive power of each predictor bundle, we evaluated the test accuracy of a multinomial logistic regression model without a given predictor bundle and compared it to the test accuracy of the full model. We then plotted the cost of each predictor bundle (direct cost, timing cost, and sum of costs) against bundle contribution to model accuracy.
Our project trajectory included many changes in analysis plan along the way. We initially attempted to incorporate longitudinal analysis and time effects into our analysis because this data is available in ADNI, and this seemed like an important aspect of Alzheimer’s prediction in a clinical setting. However, the amount of missing data associated with loss to follow up and the difficulty in dealing with censored data caused this analysis to become prohibitive. We nonetheless were able to feature engineer a delta measure of the change in estimated whole brain volume between baseline visits and 24-month visits for a majority of ADNI2 patients. This engineered feature was not included in our final models, but it would be of interest in further study.
We initially included the Clinical Dementia Rating (CDR) as a predictor in our models but found that its inclusion resulted in very high model accuracies. We believed that these accuracies were artificially inflated due to the CDR’s direct contribution to Alzheimer’s diagnosis in some cases. We therefore subsequently removed CDR from our models.
5. Results, Conclusions, Future Work:
The model classification accuracies on the training and validation sets are presented in the notebook for each of the six baseline models. As expected, each validation set accuracy is slightly lower than its corresponding training set accuracy. The classification accuracy of the multinomial logistic regression model with the full predictor set was 0.855 on the training set and 0.830 on the validation set. Finally, the accuracy of this model on the test set was 0.754, and this can be considered the most accurate measure of the final model performance.
In assessing the contribution of each predictor bundle to the overall model accuracy, we found a range of accuracy changes. Removal of both the ECogPT and MMSE from the full model, respectively, actually surprisingly resulted in slight increase of the classification accuracy. Removal of all other predictor bundles from the full model resulted in decreases in model accuracy, to varying degrees.
The figure above shows the results of plotting estimated predictor bundle cost against each bundle’s contribution to the model accuracy. Immediately obvious is that PET scan is a far more expensive test than any of the others in the analysis. It does not, however, seem to contribute the greatest amount toward predictive value. ADAS is a neuropsychological test that appears relatively cheap and increases model accuracy by > 0.03.
Stages of detection (1=early, 2=medium, 3=late) were plotted here against bundle contribution to model accuracy. Earlier detection results in cost savings as incorporated into the final figure below. In the above figure, we see that genetic testing stands out as the only early predictor, and its inclusion in the model results in model accuracy improvement of 0.025, which is relatively high compared to the best predictor bundles.
This figure incorporates the information in the above two figures by adding direct test costs to indirect costs due to timing of detection, as described in Literature Review and Modeling Approaches above. We see here an interesting spread in the summary costs by bundle importance. Because of the large estimate of monetary savings from early detection, genetics stands out as the lowest overall cost test, and its contribution to model accuracy is relatively high. ADAS improves the model the most but has a much larger associated cost, mostly due to its late detection of symptoms. Imaging modalities (MRI and PET) are the most expensive tests and notably do not result in the best predictive powers.
The strengths of our analysis include the incorporation of CSF measurements in the analysis, inclusion of a maximal number of observations from the ADNI-2 protocol via imputation of missing data, and exploration of predictor costs as related to contribution to the predictive model. Limitations of our analysis include non-perfect prediction accuracies, lack of longitudinal analysis, and inability to combine data across ADNI protocols. We are further constrained in our cost analyses by crude estimates from the literature that represent only average costs in a dynamic healthcare environment.
Future investigation could take multiple paths. Incorporation of time effects such as the difference in brain volume as measured on repeat MRI between visits would be an obvious path as we have already begun this analysis. It would be interesting to see how much time effects can contribute to model predictive power. This would also raise the question of how to penalize time effect predictors, as these measurements have an additional aspect of cost associated with them. Another possible path for future analysis is to increase the feature set either by including additional raw datasets from the ADNI project or by feature engineering to include polynomial terms and interaction terms. This would increase the need for subsequent dimensionality reduction via automated feature selection, regularization, or PCA.